More Information
Submitted: August 17, 2022 | Approved: August 26, 2022 | Published: August 29, 2022
How to cite this article: Rukadikar AR, Mendiratta DK, Siddiqui R. Identification, characterization of candida species isolated from cases of vulvovaginal candidiasis along with their antifungal susceptibility by vitek-2 system. Arch Pathol Clin Res. 2022; 6: 013-026.
DOI: 10.29328/journal.apcr.1001031
Copyright License: © 2022 Rukadikar AR, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Antifungal; Candida; Susceptibility; Vulvovaginal candidiasis; Vitek- 2 system
Identification, characterization of candida species isolated from cases of vulvovaginal candidiasis along with their antifungal susceptibility by vitek-2 system
Atul R Rukadikar1*, DK Mendiratta2 and Rafat Siddiqui3
1Associate Professor, Department of Microbiology, All India Institute of Medical Sciences, Gorakhpur, Uttar Pradesh, India
2Professor, Department of Microbiology, Jawaharlal Nehru Medical College, Sawangi (Meghe)Wardha, Maharashtra, India
3Assistant Professor, Department of Microbiology, Mahaveer Institute of Medical Sciences and Research, Bhopal, Madhya Pradesh, India
*Address for Correspondence: Dr. Atul R Rukadikar, Associate Professor, Department of Microbiology, All India Institute of Medical Sciences, Gorakhpur, Uttar Pradesh, India, Email: [email protected]
One of the most severe threats to world health is the Candida species. Many non-Candida species are the major cause of vulvovaginal candidiasis (VVC). During the development of VVC, the host environment and Candida vaginal colonization are assumed to be out of balance, and this might be owing to physiological or non-physiological changes. Host-related and behavioral risks have been connected to VVC. Novel antifungal medications with particular molecular targets may be developed with the use of molecular tools in epidemiological research and the study of resistant Candida species. Using the Vitek-2 Antifungal Susceptibility System, this review will explain the many approaches used to identify and characterize Candida species isolated from vulvovaginal candidiasis patients.
Genus Candida has a vast range of species that may be found in nature and a variety of settings, including hospitals as well as people, and domestic and wild animals [1]. These yeasts can colonize the mucosal surfaces of respiratory, vaginal, and urinary digestive systems, and the nails, scalp, and mouth cavity [2]. Non-symptomatic colonization may become infected when Candida species take over, as opposed to commensal species. They are described as opportunistic and may vary from innocuous to harmful depending on the host’s environment. However, in immunocompromised individuals, significant systemic Candida infections may arise [3]. Candida organisms enter the lower genital system mostly via the perianal region, which is close to the genital tract [4]. The delicate balance between Candida organisms and vaginal defense systems may be maintained by vaginal commensals such as lactobacilli and immunological responses, but a change in the host vaginal/vulvar microenvironment can lead to vulvovaginal candidiasis [5].
A mutation in a gene or other modification of the drug target is often to blame for Candida species’ resistance to antifungal medicines [6]. It has therefore been possible to identify and characterize Candida species using numerous viable molecular approaches. Particularly in the case of Candida species with a high level of resistance, these approaches are helpful in the development of new antifungals and epidemiological research with particular molecular targets. An overview of current knowledge on Candida species identification and characterization is presented, along with predictions for the future.
General features: candida species
These yeasts are ovoid or spherical, pleomorphic, unicellular microorganisms with an incomplete sexual cycle. Candida species may be isolated from soil, plants, water, and other habitats. They are found commensally in healthy people’s skin, vaginal mucosa, oral cavity, respiratory tracts, and gastrointestinal tracts [7]. Additionally, they may break down proteins and carbohydrates to get the carbon and nitrogen they need to grow [8].
Pathological, physiological, mechanical, and iatrogenic variables may all have an impact on how an autochthonous Candida species interacts with its human host. As a consequence, Candida species may cause a wide spectrum of infections with a wide range of clinical presentations, from superficial benign forms to invasive ones that harm several organs and cause death in the host [9]. Nearly 200 different Candida species exist, of which five - C. tropicalis, C. albicans, C. glabrata, C. parapsilosis, and C. krusei are responsible for more than 90% of invasive infections [10-12].
Individuals with superficial as well as intrusive infections of diverse anatomical locations are most often found to have C. albicans in case of studies from throughout the world [13]. Several of its most prominent pathogenic mechanisms include proteinases and phospholipases, as well as its capacity to adhere to various mucosal membranes and epithelia and create filamentous structures that aid tissue penetration. Such organisms are inherently susceptible to antifungal medications when administered systemically, however, long-term antifungal treatment patients have been shown to develop acquired resistance to azoles, particularly fluconazole [14].
There is 20% to 24% of blood-borne infections in Latin American countries, where Candida tropicalis is present, notably in elderly patients and those suffering from neutropenia and diabetes mellitus [15,16]. Amphotericin B and triazoles are usually effective against clinical isolates of this species [17], however, drug resistance to these drugs, notably fluconazole, has recently been found and described [18].
Additionally, the species C. krusei and C. glabrata have the potential to be pathogenic [15]. The second most common blood-borne infection isolates in the US is C. glabrata. There has been a rise in fluconazole-resistant isolates, reduced amphotericin B sensitivity isolates, and cross-resistance in this species to other azole-class medicines [14]. It has been shown that C. krusei may sometimes be a hospital pathogen, especially in patients who are receiving bone marrow transplants or who have hematological malignancies. Flu-conazole is inherently toxic to C. krusei [19].
From medical research and parenteral feeding solutions, C. parapsilosis has been identified [20]. This yeast has been identified as a significant contributor to skin-related infections that result in candidemia [21]. This species’ clinical isolates are often susceptible to all antifungal medications, notably azoles and amphotericin B. However, isolates with decreased sensitivity to fluconazole have been observed in clinical investigations [22]. As a group, the Candida species have considerable clinical significance because of their capacity to colonize and infect the body of humans. As a result, the kind of yeast causing an infection is to be precisely identified and diagnosed before the right antifungal medicine must be prescribed. Phenomenological techniques for the identification and categorization of Candida species rely on the physical and biochemical characteristics of these organisms. Testing for enzyme activity, assimilation capability, and substrate fermentation are some of the most popular phenotypic approaches used to identify and characterize Candida species [23].
For the diagnosis of Candida species infections, many commercial products and techniques have been developed by clinical microbiologists [24]. Among them are semi-automated and fully automated agar plates containing chromogens, kits, and panels for presumptive or definite identification of the most common species [25,26]. Because certain species of Candida have minimal morphological and biochemical changes, using simply phenotypic testing is not very efficient for identifying these species. Phenotypic and molecular approaches may be used to boost the accuracy of this identification.
Virulence factors of candida
It was formerly thought that only an organic weakness or an immunocompromised host could initiate an opportunistic fungal infection and that Candida germs passively assisted in this process. Virulence factors, as they’re often called, are processes those yeasts employ to become more aggressive and contribute to disease etiology [27].
Adhesion
Candida must adhere to host surfaces to first colonize human tissues, as it also helps the microbe stay alive within the host and is crucial for the development of infection [28]. Studies have shown that some species of Candida may cling to these cells. Additionally, some species of Candida may stick to the medical device’s surface, often encouraging infections linked to such devices, such as Vulvovaginal candidiasis in women using intrauterine devices (IUDs) as a means of contraception [29]. Candida cells first adhere to both biotic and abiotic surfaces by the action of adhesins, which are specialized cell-surface proteins that facilitate this process [30]. Laminin, fibronectin, collagen, vitronectin, and entactin are examples of extracellular matrix proteins and components that are recognized by adhesins as host ligands. Hydrophobic interactions are also used to facilitate adhesion to abiotic surfaces [31]. An ALS gene family of C. albicans has eight members (ALS1–7), which encode a considerable number of adhesins (ALS1–9). For women with VVC, Cheng, et al. discovered that all seven ALS genes were expressed in the vaginal tissues, but that only four of those genes were expressed more often than the other seven.
C. albicans ALS gene expression is also regulated by variables specific to the host site, as shown by this study group. All of the ALS genes were detected in the C. albicans-infected RHVE, with ALS3 and ALS6 displaying the highest expression and ALS7 showing the lowest expression. RHVE co-infection with C. albicans and C. glabrata reduced the expression of EPA genes, but the ALS genes were virtually the same except for an increased expression of ALS3, which was not evident in a single infection [32,33].
Development of biofilms
Accumulations of biofilms are thought to begin with Candida cells attaching themselves to surfaces such as skin or medical devices. A biofilm is a bacterial population that adheres permanently to a surface and has its extracellular matrix [34]. Biofilm communities are the most frequent form of microbial growth, accounting for up to 80% of all microorganisms found in the natural world. Microbial biofilms are also thought to have a role in more than half of all human infections. For example, biofilm-grown fungi are more resistant to antifungal medications, the host’s defense mechanisms, and physical and chemical stress than planktonic counterparts. Furthermore, biofilm cells show cooperative metabolism, community-based gene regulation, and the ability to withstand the pressure of competition from other species [35]. Because of the protection provided by biofilms, bacteria are better able to survive in severe environments. Antifungal treatment has been discovered to be up to 1000 times more effective on biofilms than on their planktonic comparable cells. This is the most important clinical property of biofilms [36]. However, the mechanisms by which Candida biofilms resist antifungal therapy are still a mystery. Multifactorial factors such as cell density, the presence of persisted cells, and stress tolerance, all contribute to biofilm resistance to antifungal drugs. In the formation of mature biofilms, environmental conditions, species, strains, and strain combinations all play a vital influence. Candida species may build biofilms on the vaginal epithelium and on IUDs, which can increase VVC in the vaginal environment [37].
Additionally, Candida infections are known to be difficult to treat because they produce biofilms in which many species coexist. While Candida biofilms that are not mono species are rare, most study is focused on Candida biofilms that are. Diagnostic and therapeutic challenges abound when dealing with mixed-species biofilms, necessitating multidrug treatment regimens of the highest order. Mixed biofilms, especially those including Candida and bacteria, may provide clinical challenges since antimicrobials designed to target a single species can promote non-targeted organisms to persist in infection. Biofilm formation on the vaginal mucosa has been shown in investigations using Candida species and vaginal bacteria like Gardenerella vaginalis, the causative agent of bacterial vaginosis (BV). However, nothing is known about how vaginal mucosal mixed Candida-bacteria biofilms arise. Women often seek medical attention due to vaginal infections, but little is known about how prevalent combined infections-in particular, VVC and BV—are. The first paper describing the frequency of BV, VVC, yeast colonization, and mixed infection was recently published by Rivers, et al. (2011). In this research, 72.5% of patients had BV diagnoses, whereas 15.7% had VVC diagnoses. 33.1% of BV-positive females had yeast colonization, and BV/VVC mixed infection was prevalent overall at 4.4%. After receiving antibiotic therapy for BV, the authors hypothesized that women may be more susceptible to VVC if they had a yeast infection or colonization. In fact, according to certain research, metronidazole or clindamycin therapy for BV often results in VVC. The outcome revealed that women with BV and yeast in their vaginal environment may either have symptom alleviation from therapy focused on one infection, or develop Vulvovaginal candidiasis from antibiotic exposure, depending on the severity of their illness. There may be little data on the prevalence of vaginal mixed infections since most illnesses in the vaginal area are diagnosed empirically rather than by using objective data. Because of the magnitude of the issue, mixed infections are probably not recognized as early as they should be, which results in ineffective treatment [39].
Extracellular hydrolytic enzyme production
Candida species release a variety of hydrolytic enzymes that are essential for adhesion, tissue penetration, invasion, and tissue annihilation [40]. Alterations in host immunological response are connected to Saps’ ability to cling to and harm host tissues. There are currently 10 SAP genes (SAP1-10) in C. albicans, three SAP genes (SAPP1-3) in C. parapsilosis, and at least four SAP genes (SAPT1-4) in C. tropicalis. However, the vast majority of NCAC species’ genes remain unidentified. Only one study has shown that C. glabrata can create proteinases; however, the kind of proteinase it makes is yet unknown. Unlike other forms of proteinases, Saps only exhibit proteinase activity at an acidic pH of 4.0. This characteristic is crucial for vulvovaginal candidiasis since the vaginal environment is acidic (pH approximately 4), creating favorable circumstances for Saps activity. According to Mohandas and Ballal (2011), who identified a relationship between Sap production and strain separation, patients with candidiasis demonstrated greater proteinase activity in vaginal isolates than in urine or respiratory isolates. According to these findings, VVC patients exhibited greater levels of SAPs expression and proteinase activity in their Candida species than carriers with no symptoms or no signs of VVC infection [41]. Additionally, it has been shown that there is a robust and precise association between vulvovaginal candidiasis and the expression of Candida albicans. In contrast to respiratory and cutaneous isolates, individuals with candidiasis had a higher concentration of phospholipase-producing strains in their vaginal isolates, according to Mohandas and Ballal (2011). The PLB and PLD gene families were also shown to be expressed in RHVE infected with a variety of Candida albicans strains, as reported by Alves, et al. (2014b). PLD1 displayed the greatest expression level, suggesting a possible function for this component in RHVE injury. Triacylglycerol hydrolysis is a step in the lipase process, which has been linked to immune cell destruction, host tissue damage, and Candida adherence. In particular, their unique relationship with the anatomical location of the infection, these enzymes are less well understood than Saps and phospholipases. The hemolysins that Candida species generate help the body regain iron, which is necessary for the organism to survive and remain within the host [42,43].
Phenotypic switching
Most Candida species have colonies that may rapidly flip between various morphologies. When it comes to NCAC species, the phenotypic flipping is far less well-understood than it is in C. albicans. Researchers say that the core of C. glabrata may change color from white to light brown or even black depending on the amount of light and darkness in the environment. Brockert, et al. (2003) found that dark brown colonies predominate in the vaginal isolates of VVC patients. Virulence attributes like adhesion, drug resistance, and Saps secretion may be affected by phenotypic switching; these changes may impact the ability of the pathogen to survive in certain anatomical areas and promote infection as a result of the changes in the pathogen’s behavior. Vaginal isolates acquired during VVC and RVVC are high-frequency modes of switching, however, the exact contribution of phenotypic switching to Vulvovaginal candidiasis is still unclear. RVVC recurrences were also discovered to have varied colony morphologies, although the DNA genotyping was always the same. Anatomical site-specific environmental variables affect every virulence component involved in Candida pathogenicity [44,45].
Identification and characterization of candida species
Genome sequencing: C. albicans SC5314 genome sequencing project began in 1996. Tools that could anticipate and identify genes in the sequence were required for this purpose. A total of 6,354 genes were identified, although the DNA sequences of certain individual chromosomes remained unknown. Genome comparisons of C. albicans and other fungus species have identified multiple genes that might be viable targets for antifungal treatments, as shown by this study. The coding sequence of the Candida albicans genome was found to be abundant in short tandem repeats compared to other fungi (STRs). Numerous multi-gene families observed in C. albicans, many of which were associated with pathogenicity, could be identified and studied in depth [46,47].
Butler, et al. [48] suggested the sequencing of Candida (WO-1), C. tropicalis (CPT-1), and Lodderomyces elongisporus (a close cousin of C. parapsilosis), as well as comparison to C. albicans SC5314 and a marine yeast infrequently linked with Candida (Debyomyces hansenii) genomes. According to Butler, et al. [48], the number of protein-coding genes in each of the species analyzed varied between 10.6 and 15.5 Mb. Candida pathogenicity has been related to 64 gene families, according to researchers. Six of these families have their pathogenicity previously related to the ERG3 gene, which is a component of the ergosterol synthesis pathway. Faster progress in figuring out the molecular mechanisms by which Candida and Non-Albicans Candida become pathogens has been made possible because of the availability of the genomes of both [49].
It has been possible to avoid the limitations of conventional methods of phenotypic identification by using molecular biology techniques for specific Candida species. MtDNA and ribosomal DNA (rDNA) analysis are the cornerstones of fungi molecular systematics [50].
Mitochondrial cytochrome C oxidase subunit 1 was identified as the molecular marker for species identification in this study (COI). The Consortium for the Barcode of Life has chosen this gene for the categorization of all organismal groupings, including fungi, some fungal genera, such as Penicillium, may be reliably identified using COI, while findings in other groups that have been experimentally tested are inconclusive [51-54].
Genes for ribosomal proteins are found in every known creature. Thus, it is possible to rebuild the phylogenies of prokaryotic and eukaryotic organisms using this gene. A low nucleotide substitution rate, however, makes it difficult to tell among closely related species in the 18S rRNA gene. Ribosomal genes, on the other hand, show that species have diverged over millions of years [55].
D1/D2 region and the Internal Transcribed Spacer regions (ITS 1 and ITS 2) have proven useful in species-level identification of closely related filamentous fungus or yeast species and subunits of rDNA for species-level identification (Figure 1). There are 100-200 tandem repeats in both ITS1 and ITS2 that comprise both highly conserved and variable regions [56]. The Consortium for the Barcode of Life has approved these areas as standard markers for use in bar-coding for the majority of fungal taxa [57]. According to Kurtzman, et al. [58], there is substantial genetic differentiation in the D1/D2 area, which allows ascomycetes to be differentiated. Using this area and the ITS regions, scientists have been able to identify many fungal species, including Candida species, and determine their evolutionary connections (phylogenies). Molecule-based phylogenies may be used to identify higher taxonomic categories, such as the Fungi kingdom, as well as the evolutionary divides between species [50].
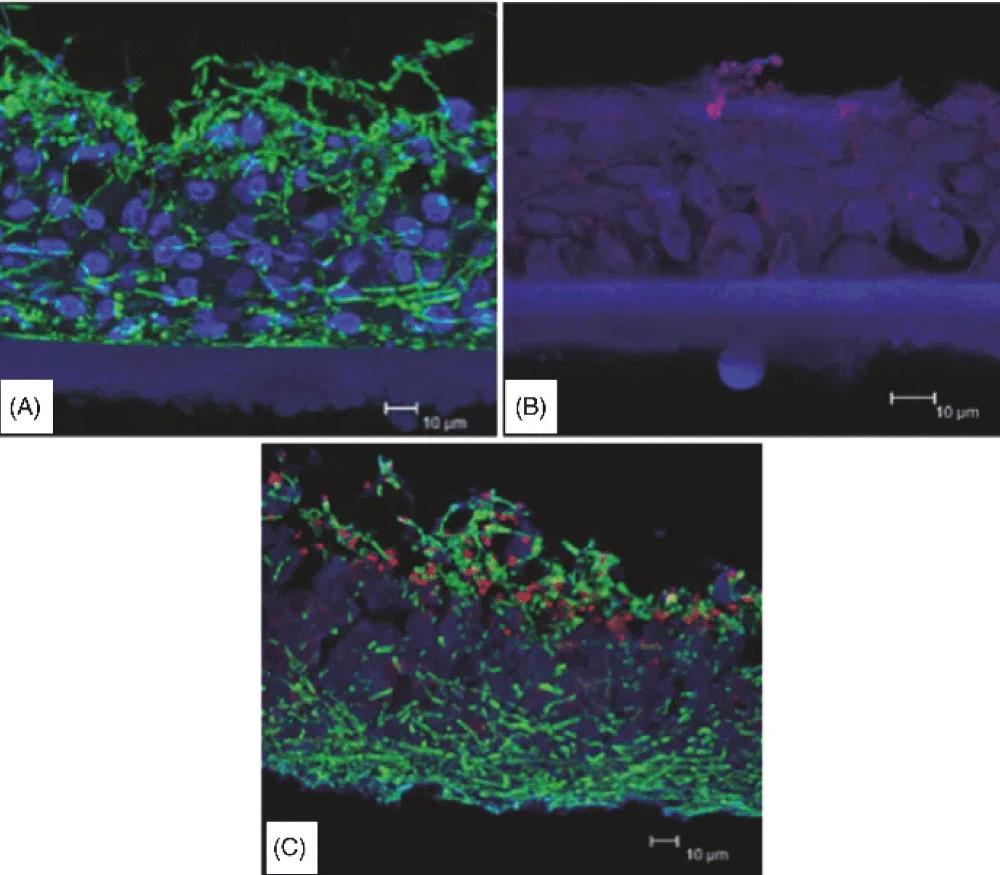
Figure 1: Reconstituted human vaginal epithelium (RHVE) infected with C. albicans (A), with C. glabrata (B) and co-infected with C. albicans and C. glabrata (C) [42].
Phylogenetic analysis: Phylogenetic tree diagrams are often used to demonstrate the evolutionary history of chemicals, animals, or both [59]. It is possible to categorize phylogenetic approaches according to the distance, character, and Bayesian inference they use. For phylogenetic reconstruction, distances must be calculated, and topology must be constructed in models that employ distances.
Microarray-based system: To better understand the molecular structure, microarrays are a common tool. In terms of strain identification, microarray-based technologies have a bright future. They may provide a high degree of specificity, sensitivity, and throughput capacity. Microarrays may be used to identify individual genes or areas, and in particular, ITSs, for molecular type-specific genotyping. As a result of ongoing genome sequencing research in pathogenic yeasts, whole-genome DNA microarray design is fairly simple [60].
Multilocus sequence typing: First, a sequencing-based technique was used to identify and identify bacterial pathogens [61]. Analysis of nucleotide polymorphisms in “housekeeping gene” fragments generates a molecular cha-racterization with high discriminating power and reliability, which can be used for a wide range of samples [62]. Many Candida species have already been studied using the MLST method. Among them is C. albicans [63,64]. Seven key C. albicans genes have been recommended for study based on joint effort [65]. It contains ACC1, AAT1a, ADP1, ZWF1b, SYA1, MPIB, and VPS13, which was later renamed PMI1 [66]. For epidemiological differentiation of C. albicans clinical isolates, MLST was demonstrated to be helpful [63,64]. In Tavanti, et al. [67], 416 C. albicans isolates from various sources were analyzed for MLST and found to have four main and eight minor clades. More C. albicans isolates (1391) were tested for MLST analysis by Odds, et al. [68], and the number of clades detected rose to 17. No correlation was discovered between antifungal susceptibility and anatomical source, however, there were statistically significant variations across clades in ABC types and geographical origins. Additionally, Wang, et al. [69] found that C. albicans mitochondrial genes are a suitable target for genotyping and population genetics.
The oral cavities and vagina of Chinese candidosis vulvovaginal patients and asymptomatic carriers were examined by Ge, et al. [70] for the presence of C. albicans. These strains’ phenotypes were uncovered. CAI 30–45 and CAI 32–46, the two major genotypes linked with vulvovaginitis, were shown to have considerably differing azole-susceptibility levels. C. albicans isolates from various genotypes and origins have varied mutation patterns in the azole target gene ERG11. Two homozygous nonsynonymous alterations in Erg11p were discovered in CAI 32–46 isolates when compared to the ERG11 sequencing of strain SC5314. Antifungal treatment may be influenced by the genotype of the C. albicans bacteria.
MLST for C. glabrata was created by Dodgson, et al. [71] by amplification and sequencing of six gene coding segments (FKS, LEU2, NMT1, TRP1, UGP1, and URA3). Two hundred and thirty-three C. glabrata isolates from five geographically and chronologically diverse populations were subjected to MLST analysis, and the results showed that the six loci may be used to evaluate genetic diversity and differentiation among the species’ isolates [72].
A study by Tavanti, et al. [73] showed that MLST can distinguish between C. tropicalis isolates with a high degree of repeatability and discriminatory power exceeding 98%. Of the 106 DST isolates examined, the technique was able to distinguish between 87 of them. MLST investigation of the C. tropicalis molecular phylogeny by Jacobsen, et al. [74] found several recombination events in their examination of the haplotypes of 242 isolates.
Only three of the 61 C. tropicalis isolates studied by Magri, et al. [75] were found to be resistant to fluconazole, although the Diploid Sequence Types (DSTs) could not be linked to resistance. For the study of genetic diversity and polymorphisms, MLST was shown to be a valuable tool by the same researchers that discovered MLST. It has become possible to molecularly characterize Candida species with great success because of recent advancements in next-generation sequencing (NGS). Antifungal resistance genes may be accurately and thoroughly genotyped using this approach. Candida isolates were studied using NGS to determine their resistance to echinocandins and antifungals. Forty isolates of Candida were examined for the presence of six antifungal resistance-related genes (ERG3, ERG11, TAC1, CgPDR1, FKS1, and FKS2) [76]. This is the first time these SNPs have been published. Both the azole and the echinocandin resistance genes were found to have new genomic changes. In a strain of C. glabrata that was resistant to echinocandins but sensitive to azoles, researchers found an FKS2 S663P mutation as well as a unique CgPDR1 mutation with a suspected loss of function. New mutations in the FKS2 gene (S663A) and a possible gain-of-function in the CgPDR1 gene were found in yet another C. glabrata strain (T370I). The results of this research reveal that next-generation sequencing (NGS) may be utilized to examine the genetics of antifungal resistance in great detail.
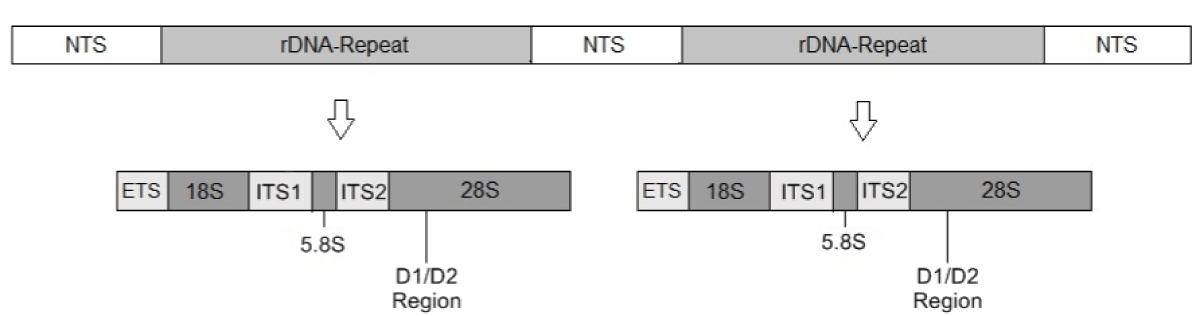
To better understand how Candida species, acquire or develop resistance to antibiotics, genome sequencing of Candida species is a useful tool. Antibiotic resistance mechanisms may be discovered using omics technologies such as the microbiome and the mycobiome as well as the genome, transcriptome, proteome, metabiome, and microbiome. This technology’s quick advancement in automation makes its eventual regular deployment more plausible as a prospect than the present low practicability and feasibility of high-throughput sequencing procedures in everyday clinical practice [77] Figure 2.
Figure 2: Structure of fungal DNA. NTS: Non- transcribed spacer, ETS: External transcribed spacer, ITS: Internal transcribed spacer, genes 18S, 5.8S and 28S ribosomal the DNA [77].
Vulvovaginal candidiasis
It is regarded to be VVC if there are no other infectious agents present, yet Candida species is present. More than one scholar has agreed on the categorization of VVC as either simple or complex depending on the level of complexity. Candida albicans cause mild to moderately severe bouts of vaginal candidiasis in otherwise healthy women who have less than four episodes per year [78].
Vaginal infections such as bacterial vaginosis, tuberculosis, trichomoniasis, and gonorrhea may all cause symptoms similar to VVC. There are many typical symptoms, including burning and itching in the vulvar area, as well as dyspareunia and dysuria. Erythema, edema, and fissures in the vulvar and vaginal areas are also prevalent. Antifungal creams, lotions, and vaginal suppositories may be used to treat VVC. Nystatin, for example, has a fungal infection cure rate of approximately 75% - 80%, whereas topical azoles (e.g., tetracycline) have a cure rate of about 85% - 90% in uncomplicated cases [79].
When compared to topical treatments, oral azole drugs (such as fluconazole and itraconazole) have equal cure rates and most patients prefer oral administration since it is more convenient. Oral azoles, on the other hand, may have hazardous adverse effects. It is far more difficult to treat more complex infections, like those caused by NCAC species or accompanied by risk factors. Preventive therapies such as topical vaginal recombinant mannose-binding lectin use and anti-Candida vaccinations use have been examined as potential preventions for VVC and anti-Candida vaccine use every year, VVC affects millions of women and is widely recognized as a significant public health issue. VVC is a significant source of mental suffering, producing pain, considerable discomfort, decreased anxiety, impaired job performance, and interfering with sexual and emotional interactions, even though it is not related to death. Furthermore, VVC has been linked to significant direct and indirect financial expenses and an increased risk of HIV infection. As a result of untreated pelvic inflammatory illness, which includes infertility, pelvic abscess, ectopic pregnancy, menstrual problems, and spontaneous abortion several consequences have been listed. As a result, preventing VVC, diagnosing it early, and treating it quickly is critical, particularly among those who are at risk [80].
Microbiology of vulvovaginal candidiasis
Candida albicans, Candida glabrata, Candida tropicalis, Candida parapsilosis, and Candida krusei are the most prevalent Candida species linked with VVC. Normally, only one species is detected, although, in certain women with VVC (1% – 10%), two or more species have been discovered [81]. Candida albicans and Candida glabrata are often found together in mixed infections [82]. Candida albicans and Candida glabrata are the two most prevalent species found in women with VVC (Table 1). An ex vivo study found that Candida glabrata penetrated vaginal tissue more readily when Candida albicans and Candida glabrata were co-infected [42].
C. albicans was historically the most common Candida species found in women with VVC, accounting for 85% – 95% of all cases [83]. However, recent studies have found incidences of C. albicans below the historical average, with some countries reporting rates as low as (Table 1). As a result of the use of antifungal therapies that were overused and improperly applied, researchers think NCAC species have become more resistant to typical antifungal drugs than C. albicans. Among patients with NCAC, RVVC species have been isolated more frequently than among those who have had sporadic VVC [84], which may be related to the fact that patients with RVVC are more likely to have been exposed to antifungals and to be using over the counter antimycotics [85]. An increase in the number of NCAC species that cause VVC, particularly C. glabrata, has been linked to advancing age [87], uncontrolled diabetes [88], and HIV infection in women [89]. In patients, hormonal imbalances and decreased immune functioning are all factors that may contribute to these correlations.
| Table 1: Summarizing the susceptibility patterns of Candida species. | ||||
| Author | Species | Anti-fungal resistance rates | Antifungal | Reference |
| Schmalreck AF, et al. 2012 | Candida. spp | 11.2% | Amphotericin B | [99] |
| Abbes S, et al. 2013; Abbes S, et al. 2014 | C. glabrata | 20.9% | Fluconazole | [42] |
| Pfaller MA, et al. 2015 | C. Albicans | 0.5% | Fluconazole | [101] |
| C. glabrata | 11.1% | Fluconazole | ||
| C. parapsilosis | 2.5% | Fluconazole | ||
| C. tropicalis | 4.5% | Fluconazole | ||
| C. guilliermondii | 20.0% | Fluconazole | ||
| C. glabrata | 1.3–2.1% | Echinocandins | ||
| C. tropicalis | 0.9–1.8% | Echinocandins | ||
| Zhang L, et al. 2015 | C. Albicans | 1.1% | Amphotericin B | [100] |
| C. krusei | 3.4% | Amphotericin B | ||
| Pfaller MA, et al. 2015 | C. glabrata | 6.8% | Fluconazole | [101] |
| Nieto MC, et al. 2015 | C. Albicans | 1.5% | Fluconazole | [102] |
| C. parapsilosis | 0.6% | Fluconazole | ||
| C. glabrata | 1.1% | Fluconazole | ||
| C. Albicans | 3.4% | Voriconazole | ||
| Wanjare S, et al. 2016 | C. Albicans | 6.67% | Caspofungin | [103] |
| Chowdhary A, et al. 2017; Calvo B, et al. 2016 | C. auris | 93% | Fluconazole | |
| 35% | Amphotericin B | |||
| 7% | Echinocandin | [104,105] | ||
According to the researcher, NCAC species are more resistant to the azoles, the most often prescribed family of antifungal drugs. NCAC species, particularly C. glabrata, which have inherently poor sensitivity to azoles and the propensity to acquire high resistance to them, have been demonstrated to produce VVC when treated with non-azole antifungals, like flucytosine and boric acid. Women with VVC who have NCAC species in their vaginas are more likely to be resistant to standard medications, which underscores the need of testing vaginal samples for these organisms to help doctors choose the best course of therapy for their patients.
Risk factors for vulvovaginal candidiasis
The vaginal microbial ecosystem is dynamic and ever-evolving. If a host’s physiological or non-physiological changes disrupt the balance between Candida vaginal colonization and the environment, yeast may thrive. Vulvovaginal candidiasis may occur at random in a healthy woman, but it is often linked to host-related and behavioral variables that modify the vaginal environment, promoting the growth and spread of the infection. Pregnancy, uncontrolled diabetic mellitus, hormone replacement medicine, immunosuppression, antibiotics, and glucocorticoid usage are among the host-related risk factors. Oral contraceptives and intrauterine devices (IUDs), spermicides, condoms, and other sexual and sanitary behaviors are all linked to an increased risk of VVC [90].
Antifungal susceptibility testing of candida species
According to several of the research previously listed and many others, Candida spp. is susceptible to in vitro infection. As a consequence of a variety of factors, it is difficult to evaluate the different outcomes. These tests may be carried out using a variety of approaches. The Clinical and Laboratory Standards Institute (CLSI) in the United States and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) in Europe have developed standardized procedures [91-93]. Methodological variations include glucose content, inoculum size, microtitration well form, and endpoint reading between the two broth dilution procedures (visual or spectrophotometric). Polyenes, azoles, and echinocandins tend to have comparable MIC values for identical isolates, with a few notable deviations [94,95]. It’s particularly important if you’re testing for resistant isolates [96]. Several “drug/bug” combos may provide a challenge to testing, such as caspofungin and C. glabrata. Furthermore, EUCAST (and Etest) studies show a trend to reduce MIC values by one to two dilution steps. Commercially accessible test instruments have also been thoroughly evaluated for their capabilities. The Etest was shown to have a strong correlation with the reference techniques [96]. The EUCAST approach categorized 2.6% of the germs as resistant in vitro, whereas the CLSI method classified 1.6% of the strains as resistant in vitro (a very significant mistake) [97]. The Vitek 2 findings and CLSI and EUCAST results vary because of discrepancies in clinical breakpoints recommended by the two organizations. However, these findings show that despite the large differences between CLSI’s CBP and EUCAST, this discrepancy only affects a small number of strains at this time.
CLSI’s “susceptible dose-dependent” (S-DD) classification of MIC data is used to categorize MIC results into S, I, and R, respectively. This has made it difficult to compare and appraise research where just percentages of S, I, and R were reported. There may be many reasons for the stated MIC50/90 values, including differing MIC distributions. As a result, MIC distributions should be used to disseminate useful surveillance data. In this way, the data may be analyzed even if, for example, the CBP had been altered, as has been the case in the past. As long as the antifungal medication in issue is designed to target a specific species, in vitro test findings would be safe in this case. To avoid arbitrary test findings, CBP should not split WT distributions. N-WT strains may be studied by monitoring the propagation of resistance mechanisms in surveillance investigations [98].
VITEK 2 fungal susceptibility test
In clinical laboratories, the spectrophotometric approach for antifungal susceptibility testing has been shown to be valid and practical [106,107] and is a key component of the EUCAST method [108,109]. Before the incorporation of spectrophotometry into commercially accessible testing methods, labs had to deal with an antifungal susceptibility testing technique that many considered difficult and time-consuming to execute [110,111]. The antifungal susceptibility test developed by bioMérieux (Hazelwood, MO) recently enables the automated testing of Candida species against four antifungal drugs (amphotericin B, fluconazole, flucytosine, and voriconazole) using the VITEK 2 microbiology system [112]. Inoculum preparation, filling of the device, incubation time and temperature, and MIC endpoint determination may all be standardized by using the fully automated VITEK 2 system, which has all the important parameters for antifungal susceptibility testing. Clinical labs will be able to conduct both fungal identification and antifungal susceptibility testing concurrently utilizing a fully automated and thoroughly standardized format if the antifungal susceptibility test is added to the VITEK 2 system [113]. VITEK 2 system MICs were shown to be in excellent agreement with reference BMD MICs for amphotericin B, fluconazole, flucytosine, and voriconazole in comparisons of Candida spp. isolates. Antifungal susceptibility testing is the first commercially accessible automated method, and this technology offers the best possible standards. To top it all off, the VITEK 2 system has a turnaround time of only 12 to 15 hours on findings that are as accurate as the reference BMDs [114,115]. Quantitative antifungal susceptibility data will play an important role in improving the management of invasive candidal infections [116].
Patients with candidemia and their Candida species identification and antifungal susceptibility testing (ID&AST) findings from January 2017 to December 2020 were studied by Andini Wulandar, et al. 2021 in a retrospective descriptive study at Dr. Kariadi Hospital, Indonesia. During these four years, the Vitek-2 system was used to determine Candida species identification and antifungal susceptibility testing. SPSS 25. Sav was used to evaluate the data once it was collected. Distributive data was examined and displayed in a table and a bar chart. A total of 85 different types of Candida were found in the bodies of the 74 people who took part in the study. The majority of candidemia cases were found in patients under the age of one year old (58.10%) or in critical care units (64.86%). Candida albicans and C. parapsilosis were the three most prevalent species isolated throughout the research period (25%). Fluconazole, Voriconazole, Caspofungin, Micafungin, and Flucytosine were all shown to be effective against 99.9% of Candida isolates, whereas Amphotericin B was found to be effective against 95.77% of Candida isolates. In 2020, a C. glabrata isolate developed resistance to Echinocandins. Non-Albicans Candida was shown to predominate in this investigation, which was performed in a tertiary care Indonesian hospital [117].
Resistance mechanism and surveillance
Candida spp. has been shown to have a variety of resistance mechanisms. The combination of multiple of these pathways may progressively lead to clinically significant resistance. Resistant fungi to fluconazole can be caused by mutations in the drug target enzyme sterol 14-demethylase, increased expression of ERG11, or overexpression of genes for membrane transport proteins from the ABC transporter (CDR1) and major facilitator families (MDR1), as well as overexpression of ERG11 [118].
The echinocandin target enzyme 1-3-b-D-glucan synthase, which is encoded by the gene fks1, was shown to be less sensitive in Candida isolates that revealed mutations in specific areas of fks1 [119]. Reduced sensitivity to echinocandins has been linked to changes in the serine at positions 645 and 641 in particular [120].
MIC values, pharmacokinetics (reported as serum AUC), and outcome [121] are all linked, at least in the case of fluconazole. Since resistance to echinocandins has been so restricted, it is impossible to establish a link between the two. Adding to the dilemma is the fact that existing antifungals do not meet the cure rates of antibacterial agents for germs that are sensitive to them. Furthermore, at least in some instances, the resistance mechanisms revealed were linked to clinical failure in the individuals implicated [122,123]. According to the CLSI, this has generated concerns about the CBP [124,125].
The ARTEMIS program is one of the world’s biggest and longest-running surveillance systems. Recent research [126] compared fluconazole and voriconazole susceptibility data for over 190,000 isolates collected between 2001 and 2007 and analyzed the resistance rates by year, geographic area, and specimen type (including humans). According to their findings, fluconazole resistance is more likely to be found in uncommon species including N. norvegensis, C. glabrata, inconspicua, guillermondii, kruseirugosa, and Famata. Pfaller, et al. reported echinocandin MIC distributions. It has been shown that echinocandin antifungals have a comparable range and efficacy for a wide range of clinically important Candida species examined. There are species with reduced susceptibilities to all three echinocandins, including C. parapsilosis and C. guilliermondii there is considerable debate about the therapeutic implications of these higher MICS values [127-131] (Table 2).
| Table 2: Agreement between results obtained by the Vitek 2 yeast susceptibility test and CLSI BMD method, with data classified by Candida species [132]. | ||||||||||
| Species (nb) | Test system | Antifungal agent | % EA | No. (%) of isolates with the result | % CA | % Error | ||||
| S | SDD | R | VME | ME | Minor | |||||
| C. glabrata | Vitek 2 | Fluconazole | 100 | 23 | 15 | 46 | 96.4 | 0 | 0 | 3.6 |
| BMD | Fluconazole | 24 | 12 | 48 | ||||||
| Vitek 2 | Voriconazole | 100 | 42 | 13 | 29 | 97.6 | 0 | 0 | 2.4 | |
| BMD | Voriconazole | 41 | 13 | 30 | ||||||
| C. Albicans | Vitek 2 | Fluconazole | 91.6 | 17 | 8 | 11 | 100 | 0 | 0 | 0 |
| BMD | Fluconazole | 17 | 8 | 11 | ||||||
| Vitek 2 | Voriconazole | 94.4 | 27 | 4 | 5 | 97.2 | 0 | 0 | 2.8 | |
The commensal status of opportunistic Candida organisms can only be changed from pathogenic to commensal by altering the vaginal environment. Finding and controlling risk factors may help prevent the spread of VVC infection. Having a predisposition to VVC infection raises your risk, but it is not a guarantee that you will get infected. Furthermore, even in the absence of a recognized risk factor, VVC may still develop. Despite progress in study, a lot of processes involved in the formation of VVC and RVVC remain obscure. We must learn more about Candida vaginal pathogenicity and its underlying processes because of the high occurrence of VVC, its unfavorable effects, and the rise in antifungal failure in its treatment. As a result of these studies, new targets for more effective treatments like the Vitek-2 system yeast susceptibility test against this clinically significant fungal infection will be discovered.
- Sidrim JJ, Rocha MF. Medical mycology in the light of contemporary authors. Guanabara Koogan; 2004.
- Anaissie EJ, Anstead GM, Arias CA, Arora A, Baer SL, Bazan III C, Bradsher Jr RW, Chintapalli KN, Deresinski SC, Diekema DJ, Dignani MC. Clinical Mycology 2nd Ed [PDF] [tahir99] VRG. pdf.
- Ascioglu S, Rex JH, De Pauw B, Bennett JE, Bille J, Crokaert F, Denning DW, Donnelly JP, Edwards JE, Erjavec Z, Fiere D. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Problems of Medical Mycology. 2003; 5(1):10-6.
- Beigi RH, Meyn LA, Moore DM, Krohn MA, Hillier SL. Vaginal yeast colonization in nonpregnant women: a longitudinal study. Obstet Gynecol. 2004 Nov;104(5 Pt 1):926-30. doi: 10.1097/01.AOG.0000140687.51048.73. PMID: 15516380.
- Ferrer J. Vaginal candidosis: epidemiological and etiological factors. Int J Gynaecol Obstet. 2000 Dec;71 Suppl 1:S21-7. doi: 10.1016/s0020-7292(00)00350-7. PMID: 11118561.
- Barker KS, Rogers PD. Recent insights into the mechanisms of antifungal resistance. Curr Infect Dis Rep. 2006 Nov;8(6):449-56. doi: 10.1007/s11908-006-0019-3. PMID: 17064638.
- Barbedo LS, Sgarbi DBG. Candidíase. J. Bras. Doenças Sex. Transm. 2010;22(1):22-38.
- Giolo MP, Svidzinski TIE. Pathophysiology, epidemiology and laboratory diagnosis of candidemia. J Bras Patol Med Lab. 2010;46(3):225- 234
- Yang YL. Virulence factors of Candida species. J Microbiol Immunol Infect. 2003;36(4):223-8
- Butler G. Fungal Sex and Pathogenesis. Clin. Microbiol. Rev. 2010;23(1):140-159.
- Colombo AL, Guimarães T. Epidemiologia das infecções hematogênicas por Candida spp [Epidemiology of hematogenous infections due to Candida spp]. Rev Soc Bras Med Trop. 2003 Sep-Oct;36(5):599-607. Portuguese. doi: 10.1590/s0037-86822003000500010. Epub 2003 Oct 21. PMID: 14576875.
- Gómez J, García-Vázquez E, Hernández A, Espinosa C, Ruiz J. Candidemias nosocomiales: nuevos retos de un problema emergente [Nosocomial candidemia: new challenges of an emergent problem]. Rev Esp Quimioter. 2010 Dec;23(4):158-68. Spanish. PMID: 21191553.
- Nguyen K, Zmeter G, Claris O, Kassai B. Epidemiology of invasive Candida infection in a neonatal intensive care unit in France. ActaPediat. 2012;101:137-139.
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007 Jan;20(1):133-63. doi: 10.1128/CMR.00029-06. PMID: 17223626; PMCID: PMC1797637.
- Nucci M, Colombo AL. Candidemia due to Candida tropicalis: clinical, epidemiologic, and microbiologic characteristics of 188 episodes occurring in tertiary care hospitals. Diagn Microbiol Infect Dis. 2007 May;58(1):77-82. doi: 10.1016/j.diagmicrobio.2006.11.009. Epub 2007 Mar 26. PMID: 17368800.
- Sipsas NV, Lewis RE, Tarrand J, Hachem R, Rolston KV, Raad II, Kontoyiannis DP. Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001-2007): stable incidence but changing epidemiology of a still frequently lethal infection. Cancer. 2009 Oct 15;115(20):4745-52. doi: 10.1002/cncr.24507. PMID: 19634156.
- Godoy P, Tiraboschi IN, Severo LC, Bustamante B, Calvo B, Almeida LP, da Matta DA, Colombo AL. Species distribution and antifungal susceptibility profile of Candida spp. bloodstream isolates from Latin American hospitals. Mem Inst Oswaldo Cruz. 2003 Apr;98(3):401-5. doi: 10.1590/s0074-02762003000300020. Epub 2003 Jul 18. PMID: 12886424.
- Almeida AA, Mesquita CS, Svidzinski TI, Oliveira KM. Antifungal susceptibility and distribution of Candida spp. isolates from the University Hospital in the municipality of Dourados, State of Mato Grosso do Sul, Brazil. Rev Soc Bras Med Trop. 2013 May-Jun;46(3):335-9. doi: 10.1590/0037-8682-0074-2012. PMID: 23856873.
- Goldman M, Pottage Júnior JC, Weaver DC. Candida kruseifungemia. Report of 4 cases and review of the literature. Medicine [Baltimore]. 1993;72(3):143-150.
- TrofaD, Gácser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Ver. 2008; 21(4):606-625.
- Brito LR, Guimarães T, Nucci M, Rosas RC, Paula Almeida L, Da Matta DA, Colombo AL. Clinical and microbiological aspects of candidemia due to Candida parapsilosis in Brazilian tertiary care hospitals. Med Mycol. 2006 May;44(3):261-6. doi: 10.1080/13693780500421476. PMID: 16702106.
- van Asbeck EC, Huang YC, Markham AN, Clemons KV, Stevens DA. Candida parapsilosis fungemia in neonates: genotyping results suggest healthcare workers hands as source, and review of published studies. Mycopathologia. 2007 Dec;164(6):287-93. doi: 10.1007/s11046-007-9054-3. Epub 2007 Sep 15. PMID: 17874281.
- Spolidorio DMP, Boriollo MFG, Carlos Estrela C, Spolidorio LC. Diferentes métodos fenotípicos para isolamento e identificação de espécies deCandida. Robrac. 2009;18(45):18-26.
- Higashi CM, Takashina FH, Rechenchoski ZD3, Stipp-Abe AT, Vespero EC, Quesada RMB, et al. Comparison of the Vitek 2 automated identification system and PCR-ITS for species characterization of clinical isolates of Candidaspp. Biological and Health Sciences. 2015; 36(1):233-242.
- Meletiadis J, Arabatzis M, Bompola M, Tsiveriotis K, Hini S, Petinaki E, Velegraki A, Zerva L. Comparative evaluation of three commercial identification systems using common and rare bloodstream yeast isolates. J Clin Microbiol. 2011 Jul;49(7):2722-7. doi: 10.1128/JCM.01253-10. Epub 2011 May 4. PMID: 21543578; PMCID: PMC3147866.
- Kumar S, Vyas A, Kumar M, Mehra SK. Application of CHROMagarCandida for identification of clinically important Candida species and their antifungal susceptibility pattern. IJBMR. 2013;4(4):3600-3606.
- Tamura NK, Negri MF, Bonassoli LA, Svidzinski TI. Fatores de virulência de Candida spp isoladas de cateteres venosos e mãos de servidores hospitalares [Virulence factors for Candida spp recovered from intravascular catheters and hospital workers hands]. Rev Soc Bras Med Trop. 2007 Jan-Feb;40(1):91-3. Portuguese. doi: 10.1590/s0037-86822007000100021. PMID: 17486265.
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012 Mar;36(2):288-305. doi: 10.1111/j.1574-6976.2011.00278.x. Epub 2011 Jun 6. PMID: 21569057.
- Ahmed A, Bashir M. Association of infection among intrauterine contraceptive device users in family planning clinics in Sudan.
- Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Mol Microbiol. 2006 Apr;60(1):5-15. doi: 10.1111/j.1365-2958.2006.05072.x. PMID: 16556216.
- Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev. 2008 Sep;72(3):495-544. doi: 10.1128/MMBR.00032-07. PMID: 18772287; PMCID: PMC2546859.
- Cheng G, Wozniak K, Wallig MA, Fidel PL Jr, Trupin SR, Hoyer LL. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun. 2005 Mar;73(3):1656-63. doi: 10.1128/IAI.73.3.1656-1663.2005. PMID: 15731066; PMCID: PMC1064955.
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009 Jun 4;459(7247):657-62. doi: 10.1038/nature08064. PMID: 19465905; PMCID: PMC2834264.
- Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003 Jan;11(1):30-6. doi: 10.1016/s0966-842x(02)00002-1. PMID: 12526852.
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002 Sep;8(9):881-90. doi: 10.3201/eid0809.020063. PMID: 12194761; PMCID: PMC2732559.
- Ramage G, Rajendran R, Sherry L, Williams C. Fungal biofilm resistance. Int J Microbiol. 2012;2012:528521. doi: 10.1155/2012/528521. Epub 2012 Feb 8. PMID: 22518145; PMCID: PMC3299327.
- Silva S, Henriques M, Martins A, Oliveira R, Williams D, Azeredo J. Biofilms of non-Candida albicans Candida species: quantification, structure and matrix composition. Med Mycol. 2009 Nov;47(7):681-9. doi: 10.3109/13693780802549594. PMID: 19888800.
- Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Noverr MC. Candida albicans forms biofilms on the vaginal mucosa. Microbiology (Reading). 2010 Dec;156(Pt 12):3635-3644. doi: 10.1099/mic.0.039354-0. Epub 2010 Aug 12. PMID: 20705667; PMCID: PMC3068702.
- Rivers CA, Adaramola OO, Schwebke JR. Prevalence of bacterial vaginosis and vulvovaginal candidiasis mixed infection in a southeastern american STD clinic. Sex Transm Dis. 2011 Jul;38(7):672-4. doi: 10.1097/OLQ.0b013e31820fc3b8. PMID: 21844715.
- Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005 Nov;48(6):365-77. doi: 10.1111/j.1439-0507.2005.01165.x. PMID: 16262871.
- Mohandas V, Ballal M. Distribution of Candida species in different clinical samples and their virulence: biofilm formation, proteinase and phospholipase production: a study on hospitalized patients in southern India. J Glob Infect Dis. 2011 Jan;3(1):4-8. doi: 10.4103/0974-777X.77288. PMID: 21572601; PMCID: PMC3068577.
- Alves CT, Wei XQ, Silva S, Azeredo J, Henriques M, Williams DW. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J Infect. 2014 Oct;69(4):396-407. doi: 10.1016/j.jinf.2014.06.002. Epub 2014 Jun 9. PMID: 24924556.
- Zarrinfar H, Kaboli S, Dolatabadi S, Mohammadi R. Rapid detection of Candida species in bronchoalveolar lavage fluid from patients with pulmonary symptoms. Braz J Microbiol. 2016 Jan-Mar;47(1):172-6. doi: 10.1016/j.bjm.2015.02.001. Epub 2016 Jan 27. PMID: 26887241; PMCID: PMC4822774.
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007 Oct;5(10):e256. doi: 10.1371/journal.pbio.0050256. PMID: 17880264; PMCID: PMC1976629.
- Brockert PJ, Lachke SA, Srikantha T, Pujol C, Galask R, Soll DR. Phenotypic switching and mating type switching of Candida glabrata at sites of colonization. Infect Immun. 2003 Dec;71(12):7109-18. doi: 10.1128/IAI.71.12.7109-7118.2003. PMID: 14638801; PMCID: PMC308932.
- Braun BR, van Het Hoog M, d'Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, Bachewich C, Harcus D, Marcil A, Dignard D, Iouk T, Zito R, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro CA, Bates S, Gow NA, Hoyer LL, Köhler G, Morschhäuser J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Costanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell AP, Johnson AD, Whiteway M, Nantel A. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005 Jul;1(1):36-57. doi: 10.1371/journal.pgen.0010001. Epub 2005 Jun 17. PMID: 16103911; PMCID: PMC1183520.
- Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, Newport G, Thorstenson YR, Agabian N, Magee PT, Davis RW, Scherer S. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004 May 11;101(19):7329-34. doi: 10.1073/pnas.0401648101. Epub 2004 May 3. PMID: 15123810; PMCID: PMC409918.
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009 Jun 4;459(7247):657-62. doi: 10.1038/nature08064. PMID: 19465905; PMCID: PMC2834264.
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuvéglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisramé A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wésolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004 Jul 1;430(6995):35-44. doi: 10.1038/nature02579. PMID: 15229592.
- Bridge PD, Spooner BM, Roberts PJ. The impact of molecular data in fungal systematics. InAdvances in botanical research. Academic Press; 2005; 42: 33-67).
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003 Feb 7;270(1512):313-21. doi: 10.1098/rspb.2002.2218. PMID: 12614582; PMCID: PMC1691236.
- Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature. 2005 May 5;435(7038):17. doi: 10.1038/435017b. PMID: 15874991.
- Seifert KA, Samson RA, Dewaard JR, Houbraken J, Lévesque CA, Moncalvo JM, Louis-Seize G, Hebert PD. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):3901-6. doi: 10.1073/pnas.0611691104. Epub 2007 Feb 28. PMID: 17360450; PMCID: PMC1805696.
- Dentinger BT, Didukh MY, Moncalvo JM. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina). PLoS One. 2011;6(9):e25081. doi: 10.1371/journal.pone.0025081. Epub 2011 Sep 22. PMID: 21966418; PMCID: PMC3178597.
- Simon L, BousquetJ, Levesque RC, Lalonde M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature. 1993;363:67-69.
- Shokohi T, Hashemi Soteh MB, Saltanat Pouri Z, Hedayati MT, Mayahi S. Identification of Candida species using PCR-RFLP in cancer patients in Iran. Indian J Med Microbiol. 2010 Apr-Jun;28(2):147-51. doi: 10.4103/0255-0857.62493. PMID: 20404462.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6241-6. doi: 10.1073/pnas.1117018109. Epub 2012 Mar 27. PMID: 22454494; PMCID: PMC3341068.
- Kurtzman CP, Albertyn J, Basehoar-Powers E. Multigene phylogenetic analysis of the Lipomycetaceae and the proposed transfer of Zygozyma species to Lipomyces and Babjevia anomala to Dipodascopsis. FEMS Yeast Res. 2007 Sep;7(6):1027-34. doi: 10.1111/j.1567-1364.2007.00246.x. Epub 2007 May 10. PMID: 17498213.
- Brinkman FS, Leipe DD. Phylogenetic analysis. Methods Biochem Anal. 2001;43:323-58. doi: 10.1002/0471223921.ch14. PMID: 11449731.
- Cummings CA, Relman DA. Using DNA microarrays to study host-microbe interactions. Emerg Infect Dis. 2000 Sep-Oct;6(5):513-25. doi: 10.3201/eid0605.000511. PMID: 10998383; PMCID: PMC2627958.
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998 Mar 17;95(6):3140-5. doi: 10.1073/pnas.95.6.3140. PMID: 9501229; PMCID: PMC19708.
- Odds FC, Jacobsen MD. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell. 2008 Jul;7(7):1075-84. doi: 10.1128/EC.00062-08. Epub 2008 May 2. PMID: 18456859; PMCID: PMC2446668.
- Bougnoux ME, Morand S, d'Enfert C. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J Clin Microbiol. 2002 Apr;40(4):1290-7. doi: 10.1128/JCM.40.4.1290-1297.2002. PMID: 11923347; PMCID: PMC140389.
- Tavanti A, Gow NA, Senesi S, Maiden MC, Odds FC. Optimization and validation of multilocus sequence typing for Candida albicans. J Clin Microbiol. 2003 Aug;41(8):3765-76. doi: 10.1128/JCM.41.8.3765-3776.2003. PMID: 12904388; PMCID: PMC179823.
- Bougnoux ME, Tavanti A, Bouchier C, Gow NA, Magnier A, Davidson AD, Maiden MC, D'Enfert C, Odds FC. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol. 2003 Nov;41(11):5265-6. doi: 10.1128/JCM.41.11.5265-5266.2003. PMID: 14605179; PMCID: PMC262540.
- Arnaud MB, Costanzo MC, Skrzypek MS, Shah P, Binkley G, Lane C, Miyasato SR, Sherlock G. Sequence resources at the Candida Genome Database. Nucleic Acids Res. 2007 Jan;35(Database issue):D452-6. doi: 10.1093/nar/gkl899. Epub 2006 Nov 7. PMID: 17090582; PMCID: PMC1669745.
- Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J Clin Microbiol. 2005 Nov;43(11):5601-13. doi: 10.1128/JCM.43.11.5601-5613.2005. PMID: 16272493; PMCID: PMC1287804.
- Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li SY, Tavanti A, Maiden MC, Gow NA, d'Enfert C. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007 Jun;6(6):1041-52. doi: 10.1128/EC.00041-07. Epub 2007 Apr 6. PMID: 17416899; PMCID: PMC1951527.
- Wang H, Guo H, Sun S, Xu J. Abundant sequence variation around the mitochondrial origin of replication in the human opportunistic yeast pathogen Candida albicans from a tropical island in China. Fungal Genet Biol. 2007 Oct;44(10):991-1001. doi: 10.1016/j.fgb.2007.03.005. Epub 2007 Apr 2. PMID: 17493848.
- Ge SH, Wan Z, Li J, Xu J, Li RY, Bai FY. Correlation between azole susceptibilities, genotypes, and ERG11 mutations in Candida albicans isolates associated with vulvovaginal candidiasis in China. Antimicrob Agents Chemother. 2010 Aug;54(8):3126-31. doi: 10.1128/AAC.00118-10. Epub 2010 Jun 1. PMID: 20516286; PMCID: PMC2916359.
- Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol. 2003 Dec;41(12):5709-17. doi: 10.1128/JCM.41.12.5709-5717.2003. PMID: 14662965; PMCID: PMC309006.
- Lott TJ, Frade JP, Lockhart SR. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. surveillance studies. Eukaryot Cell. 2010 Apr;9(4):619-25. doi: 10.1128/EC.00002-10. Epub 2010 Feb 26. PMID: 20190071; PMCID: PMC2863408.
- Tavanti A, Davidson AD, Johnson EM, Maiden MC, Shaw DJ, Gow NA, Odds FC. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J Clin Microbiol. 2005 Nov;43(11):5593-600. doi: 10.1128/JCM.43.11.5593-5600.2005. PMID: 16272492; PMCID: PMC1287820.
- Jacobsen MD, Rattray AM, Gow NA, Odds FC, Shaw DJ. Mitochondrial haplotypes and recombination in Candida albicans. Med Mycol. 2008 Nov;46(7):647-54. doi: 10.1080/13693780801986631. PMID: 18608923.
- Magri MM, Gomes-Gouvêa MS, de Freitas VL, Motta AL, Moretti ML, Shikanai-Yasuda MA. Multilocus sequence typing of Candida tropicalis shows the presence of different clonal clusters and fluconazole susceptibility profiles in sequential isolates from candidemia patients in Sao Paulo, Brazil. J Clin Microbiol. 2013 Jan;51(1):268-77. doi: 10.1128/JCM.02366-12. Epub 2012 Nov 14. PMID: 23152555; PMCID: PMC3536249.
- Garnaud C, Botterel F, Sertour N, Bougnoux ME, Dannaoui E, Larrat S, Hennequin C, Guinea J, Cornet M, Maubon D. Next-generation sequencing offers new insights into the resistance of Candida spp. to echinocandins and azoles. J Antimicrob Chemother. 2015 Sep;70(9):2556-65. doi: 10.1093/jac/dkv139. Epub 2015 May 27. PMID: 26017039.
- dos Santos Silva DB, de Oliveira KM, Grisolia AB. Molecular Methods Developed for the Identification and Characterization of Candida Species.
- Awari A. Species distribution and antifungal susceptibility profile of Candida isolated from urine samples. Int J App Basic Med Res. 2011; 18:228-34.
- Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr, Calandra TF, Edwards JE Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD; Infectious Diseases Society of America. Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009 Mar 1;48(5):503-35. doi: 10.1086/596757. PMID: 19191635; PMCID: PMC7294538.
- Sobel JD. Genital candidiasis. Medicine. 2005 Oct 1;33(10):62-5.
- Nwadioha SI, Egah DZ, Alao OO, Iheanacho E. Risk factors for vaginal candidiasis among women attending primary health care centers of Jos, Nigeria. J Clin Med Res. 2010 Jul 31;2(7):110-3.
- Amouri I, Sellami H, Borji N, Abbes S, Sellami A, Cheikhrouhou F, Maazoun L, Khaled S, Khrouf S, Boujelben Y, Ayadi A. Epidemiological survey of vulvovaginal candidosis in Sfax, Tunisia. Mycoses. 2011 Sep;54(5):e499-505. doi: 10.1111/j.1439-0507.2010.01965.x. Epub 2010 Oct 29. PMID: 21039942.
- Mahmoudi Rad M, Zafarghandi S, Abbasabadi B, Tavallaee M. The epidemiology of Candida species associated with vulvovaginal candidiasis in an Iranian patient population. Eur J Obstet Gynecol Reprod Biol. 2011 Apr;155(2):199-203. doi: 10.1016/j.ejogrb.2010.11.022. Epub 2010 Dec 30. PMID: 21194828.
- Linhares LM, Witkin SS, Miranda SD, Fonseca AM, Pinotti JA, Ledger WJ. Differentiation between women with vulvovaginal symptoms who are positive or negative for Candida species by culture. Infect Dis Obstet Gynecol. 2001;9(4):221-5. doi: 10.1155/S1064744901000369. PMID: 11916179; PMCID: PMC1784657.
- Grigoriou O, Baka S, Makrakis E, Hassiakos D, Kapparos G, Kouskouni E. Prevalence of clinical vaginal candidiasis in a university hospital and possible risk factors. Eur J Obstet Gynecol Reprod Biol. 2006 May 1;126(1):121-5. doi: 10.1016/j.ejogrb.2005.09.015. Epub 2005 Oct 26. PMID: 16256258.
- Fan SR, Liao QP, Liu XP, Liu ZH, Zhang D. Vaginal allergic response in women with vulvovaginal candidiasis. Int J Gynaecol Obstet. 2008 Apr;101(1):27-30. doi: 10.1016/j.ijgo.2007.08.024. Epub 2008 Feb 15. PMID: 18280478.
- Goswami D, Goswami R, Banerjee U, Dadhwal V, Miglani S, Lattif AA, Kochupillai N. Pattern of Candida species isolated from patients with diabetes mellitus and vulvovaginal candidiasis and their response to single dose oral fluconazole therapy. J Infect. 2006 Feb;52(2):111-7. doi: 10.1016/j.jinf.2005.03.005. PMID: 15908007.
- Richter SS, Galask RP, Messer SA, Hollis RJ, Diekema DJ, Pfaller MA. Antifungal susceptibilities of Candida species causing vulvovaginitis and epidemiology of recurrent cases. J Clin Microbiol. 2005 May;43(5):2155-62. doi: 10.1128/JCM.43.5.2155-2162.2005. PMID: 15872235; PMCID: PMC1153777.
- Sobel JD, Chaim W, Nagappan V, Leaman D. Treatment of vaginitis caused by Candida glabrata: use of topical boric acid and flucytosine. Am J Obstet Gynecol. 2003 Nov;189(5):1297-300. doi: 10.1067/s0002-9378(03)00726-9. PMID: 14634557.
- Safdar A, Armstrong D, Cross EW, Perlin DS. Prospective epidemiologic analysis of triazole-resistant nosocomial Candida glabrata isolated from patients at a comprehensive cancer center. Int J Infect Dis. 2002 Sep;6(3):198-201. doi: 10.1016/s1201-9712(02)90111-6. PMID: 12718835.
- Patel DA, Gillespie B, Sobel JD, Leaman D, Nyirjesy P, Weitz MV, Foxman B. Risk factors for recurrent vulvovaginal candidiasis in women receiving maintenance antifungal therapy: results of a prospective cohort study. Am J Obstet Gynecol. 2004 Mar;190(3):644-53. doi: 10.1016/j.ajog.2003.11.027. PMID: 15041994.
- M27-a3, Reference Method for Broth dilution antifungal susceptibility testing of yeasts; approved standard - third Edition 2008, clinical and laboratory standards Institute, Wayne, usa
- Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin Microbiol Infect. 2008 Apr;14(4):398-405. doi: 10.1111/j.1469-0691.2007.01935.x. Epub 2008 Jan 11. Erratum in: Clin Microbiol Infect. 2009 Jan;15(1):103. PMID: 18190574.
- Cuenca-Estrella M, Lee-Yang W, Ciblak MA, Arthington-Skaggs BA, Mellado E, Warnock DW, Rodriguez-Tudela JL. Comparative evaluation of NCCLS M27-A and EUCAST broth microdilution procedures for antifungal susceptibility testing of candida species. Antimicrob Agents Chemother. 2002 Nov;46(11):3644-7. doi: 10.1128/AAC.46.11.3644-3647.2002. PMID: 12384382; PMCID: PMC128746.
- Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Moet GJ, Jones RN. Comparison of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Etest methods with the CLSI broth microdilution method for echinocandin susceptibility testing of Candida species. J Clin Microbiol. 2010 May;48(5):1592-9. doi: 10.1128/JCM.02445-09. Epub 2010 Mar 24. PMID: 20335424; PMCID: PMC2863935.
- Arendrup MC. Candida and candidaemia. Susceptibility and epidemiology. Dan Med J. 2013 Nov;60(11):B4698. PMID: 24192246.
- Cuenca-Estrella M, Gomez-Lopez A, Alastruey-Izquierdo A, Bernal-Martinez L, Cuesta I, Buitrago MJ, Rodriguez-Tudela JL. Comparison of the Vitek 2 antifungal susceptibility system with the clinical and laboratory standards institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) Broth Microdilution Reference Methods and with the Sensititre YeastOne and Etest techniques for in vitro detection of antifungal resistance in yeast isolates. J Clin Microbiol. 2010 May;48(5):1782-6. doi: 10.1128/JCM.02316-09. Epub 2010 Mar 10. PMID: 20220169; PMCID: PMC2863906.
- Arendrup MC, Kahlmeter G, Rodriguez-Tudela JL, Donnelly JP. Breakpoints for susceptibility testing should not divide wild-type distributions of important target species. Antimicrob Agents Chemother. 2009 Apr;53(4):1628-9. doi: 10.1128/AAC.01624-08. Epub 2009 Feb 2. PMID: 19188378; PMCID: PMC2663114.
- Schmalreck AF, Willinger B, Haase G, Blum G, Lass-Flörl C, Fegeler W, Becker K; Antifungal Susceptibility Testing-AFST Study Group. Species and susceptibility distribution of 1062 clinical yeast isolates to azoles, echinocandins, flucytosine and amphotericin B from a multi-centre study. Mycoses. 2012 May;55(3):e124-37. doi: 10.1111/j.1439-0507.2011.02165.x. Epub 2012 Jan 11. PMID: 22233267.
- Zhang L, Zhou S, Pan A, Li J, Liu B. Surveillance of antifungal susceptibilities in clinical isolates of Candida species at 36 hospitals in China from 2009 to 2013. Int J Infect Dis. 2015 Apr;33:1-4. doi: 10.1016/j.ijid.2014.12.033. Epub 2014 Dec 23. PMID: 25541294.
- Pfaller MA, Rhomberg PR, Messer SA, Castanheira M. in vitro activity of a Hos2 deacetylase inhibitor, MGCD290, in combination with echinocandins against echinocandin-resistant Candida species. Diagn Microbiol Infect Dis. 2015 Apr;81(4):259-63. doi: 10.1016/j.diagmicrobio.2014.11.008. Epub 2014 Nov 25. PMID: 25600842.
- Nieto MC, Tellería O, Cisterna R. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagn Microbiol Infect Dis. 2015 Jan;81(1):34-40. doi: 10.1016/j.diagmicrobio.2014.05.021. Epub 2014 Jun 5. PMID: 25439581.
- Wanjare S, Gupta R, Mehta P. Caspofungin MIC Distribution amongst Commonly Isolated Candida Species in a Tertiary Care Centre - An Indian Experience. J Clin Diagn Res. 2016 Nov;10(11):DC11-DC13. doi: 10.7860/JCDR/2016/23731.8883. Epub 2016 Nov 1. PMID: 28050365; PMCID: PMC5198318.
- Chowdhary A, Rhandhawa HS, Prakash A, Meis JF. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: an update. Crit Rev Microbiol. 2012 Feb;38(1):1-16. doi: 10.3109/1040841X.2011.606426. Epub 2011 Dec 1. PMID: 22133016.
- Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. First report of Candida auris in America: Clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 2016 Oct;73(4):369-74. doi: 10.1016/j.jinf.2016.07.008. Epub 2016 Jul 21. PMID: 27452195.
- Cuenca-Estrella M, Lee-Yang W, Ciblak MA, Arthington-Skaggs BA, Mellado E, Warnock DW, Rodriguez-Tudela JL. Comparative evaluation of NCCLS M27-A and EUCAST broth microdilution procedures for antifungal susceptibility testing of candida species. Antimicrob Agents Chemother. 2002 Nov;46(11):3644-7. doi: 10.1128/AAC.46.11.3644-3647.2002. PMID: 12384382; PMCID: PMC128746.
- Espinel-Ingroff A, Barchiesi F, Cuenca-Estrella M, Fothergill A, Pfaller MA, Rinaldi M, Rodriguez-Tudela JL, Verweij PE. Comparison of visual 24-hour and spectrophotometric 48-hour MICs to CLSI reference microdilution MICs of fluconazole, itraconazole, posaconazole, and voriconazole for Candida spp.: a collaborative study. J Clin Microbiol. 2005 Sep;43(9):4535-40. doi: 10.1128/JCM.43.9.4535-4540.2005. PMID: 16145103; PMCID: PMC1234107.
- Cuenca-Estrella M, Moore CB, Barchiesi F, Bille J, Chryssanthou E, Denning DW, Donnelly JP, Dromer F, Dupont B, Rex JH, Richardson MD, Sancak B, Verweij PE, Rodríguez-Tudela JL; AFST Subcommittee of the European Committee on Antimicrobial Susceptibility Testing. Multicenter evaluation of the reproducibility of the proposed antifungal susceptibility testing method for fermentative yeasts of the Antifungal Susceptibility Testing Subcommittee of the European Committee on Antimicrobial Susceptibility Testing (AFST-EUCAST). Clin Microbiol Infect. 2003 Jun;9(6):467-74. doi: 10.1046/j.1469-0691.2003.00592.x. PMID: 12848721.
- Rodriguez-Tudela JL, Barchiesi F, Bille J, Chryssanthou E, Cuenca-Estrella M, Denning D, Donnelly JP, Dupont B, Fegeler W, Moore C, Richardson M. Method for the determination of minimum inhibitory concentration (MIC) by broth dilution of fermentative yeasts. Clinical Microbiology and Infection. 2003; 9(8):i-viii.
- Forrest G. Role of antifungal susceptibility testing in patient management. Curr Opin Infect Dis 2006, 19: 538– 543.
- Pfaller MA. Antifungal susceptibility testing methods. Curr Drug Targets 2005, 6: 929– 943.
- Pfaller MA. New developments in the antifungal susceptibility testing of Candida. Current Fungal Infection Reports. 2008; Sep; 2(3):125-33.
- Aubertine CL, Rivera M, Rohan SM, Larone DH. Comparative study of the new colorimetric VITEK 2 yeast identification card versus the older fluorometric card and of CHROMagar Candida as a source medium with the new card. J Clin Microbiol. 2006 Jan;44(1):227-8. doi: 10.1128/JCM.44.1.227-228.2006. PMID: 16390976; PMCID: PMC1351943.
- Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 yeast susceptibility test with the CLSI broth microdilution reference method for testing fluconazole against Candida spp. J Clin Microbiol. 2007 Mar;45(3):796-802. doi: 10.1128/JCM.01986-06. Epub 2007 Jan 10. PMID: 17215344; PMCID: PMC1829143.
- Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine, and voriconazole against Candida spp. J Clin Microbiol. 2007 Nov;45(11):3522-8. doi: 10.1128/JCM.00403-07. Epub 2007 Oct 3. PMID: 17913927; PMCID: PMC2168477.
- Collins CD, Eschenauer GA, Salo SL, Newton DW. To test or not to test: a cost minimization analysis of susceptibility testing for patients with documented Candida glabrata fungemias. J Clin Microbiol. 2007 Jun;45(6):1884-8. doi: 10.1128/JCM.00192-07. Epub 2007 Apr 4. PMID: 17409208; PMCID: PMC1933067.
- Wulandari A, Hapsari R, Sari D, Puspitasari I, Pramukarso DT. Antifungal susceptibility profile of Candida spp. causing candidemia in an Indonesian tertiary hospital. Journal of Clinical Microbiology and Infectious Diseases. 2021; 1(2):28-32.
- Morschhäuser J. The genetic basis of fluconazole resistance development in Candida albicans. Biochim Biophys Acta. 2002 Jul 18;1587(2-3):240-8. doi: 10.1016/s0925-4439(02)00087-x. PMID: 12084466.
- Park S, Kelly R, Kahn JN, Robles J, Hsu MJ, Register E, Li W, Vyas V, Fan H, Abruzzo G, Flattery A, Gill C, Chrebet G, Parent SA, Kurtz M, Teppler H, Douglas CM, Perlin DS. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother. 2005 Aug;49(8):3264-73. doi: 10.1128/AAC.49.8.3264-3273.2005. PMID: 16048935; PMCID: PMC1196231.
- Balashov SV, Park S, Perlin DS. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother. 2006 Jun;50(6):2058-63. doi: 10.1128/AAC.01653-05. PMID: 16723566; PMCID: PMC1479158.
- Pai MP, Turpin RS, Garey KW. Association of fluconazole area under the concentration-time curve/MIC and dose/MIC ratios with mortality in nonneutropenic patients with candidemia. Antimicrob Agents Chemother. 2007 Jan;51(1):35-9. doi: 10.1128/AAC.00474-06. Epub 2006 Nov 13. PMID: 17101684; PMCID: PMC1797664.
- Hernandez S, López-Ribot JL, Najvar LK, McCarthy DI, Bocanegra R, Graybill JR. Caspofungin resistance in Candida albicans: correlating clinical outcome with laboratory susceptibility testing of three isogenic isolates serially obtained from a patient with progressive Candida esophagitis. Antimicrob Agents Chemother. 2004 Apr;48(4):1382-3. doi: 10.1128/AAC.48.4.1382-1383.2004. PMID: 15047549; PMCID: PMC375251.
- Krogh-Madsen M, Arendrup MC, Heslet L, Knudsen JD. Amphotericin B and caspofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin Infect Dis. 2006 Apr 1;42(7):938-44. doi: 10.1086/500939. Epub 2006 Feb 27. PMID: 16511756.
- Arendrup MC, Cuenca-Estrella M, Donnelly JP, Lass-Flörl C, Rodriguez-Tudela JL. Association of fluconazole pharmacodynamics with mortality in patients with candidemia. Antimicrob Agents Chemother. 2009 Jun;53(6):2704-5; author reply 2705-6. doi: 10.1128/AAC.01411-08. PMID: 19458347; PMCID: PMC2687210.
- Arendrup MC, Cuenca-Estrella M, Donnelly JP, Lass-Flörl C, Rodriguez-Tudela JL. Association of fluconazole pharmacodynamics with mortality in patients with candidemia. Antimicrob Agents Chemother. 2009 Jun;53(6):2704-5; author reply 2705-6. doi: 10.1128/AAC.01411-08. PMID: 19458347; PMCID: PMC2687210.
- Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA; Global Antifungal Surveillance Group. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010 Apr;48(4):1366-77. doi: 10.1128/JCM.02117-09. Epub 2010 Feb 17. PMID: 20164282; PMCID: PMC2849609.
- Messer SA, Moet GJ, Kirby JT, Jones RN. Activity of contemporary antifungal agents, including the novel echinocandin anidulafungin, tested against Candida spp., Cryptococcus spp., and Aspergillus spp.: report from the SENTRY Antimicrobial Surveillance Program (2006 to 2007). J Clin Microbiol. 2009 Jun;47(6):1942-6. doi: 10.1128/JCM.02434-08. Epub 2009 Apr 22. PMID: 19386851; PMCID: PMC2691076.
- Messer SA, Jones RN, Moet GJ, Kirby JT, Castanheira M. Potency of anidulafungin compared to nine other antifungal agents tested against Candida spp., Cryptococcus spp., and Aspergillus spp.: results from the global SENTRY Antimicrobial Surveillance Program (2008). J Clin Microbiol. 2010 Aug;48(8):2984-7. doi: 10.1128/JCM.00328-10. Epub 2010 Jun 9. PMID: 20534798; PMCID: PMC2916586.
- Fleck R, Dietz A, Hof H. in vitro susceptibility of Candida species to five antifungal agents in a German university hospital assessed by the reference broth microdilution method and Etest. J Antimicrob Chemother. 2007 Apr;59(4):767-71. doi: 10.1093/jac/dkl555. Epub 2007 Feb 9. PMID: 17293369.
- Diekema DJ, Messer SA, Boyken LB, Hollis RJ, Kroeger J, Tendolkar S, Pfaller MA. in vitro activity of seven systemically active antifungal agents against a large global collection of rare Candida species as determined by CLSI broth microdilution methods. J Clin Microbiol. 2009 Oct;47(10):3170-7. doi: 10.1128/JCM.00942-09. Epub 2009 Aug 26. PMID: 19710283; PMCID: PMC2756931.
- Chen SC, Marriott D, Playford EG, Nguyen Q, Ellis D, Meyer W, Sorrell TC, Slavin M; Australian Candidaemia Study. Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin Microbiol Infect. 2009 Jul;15(7):662-9. doi: 10.1111/j.1469-0691.2009.02821.x. Epub 2009 Jul 15. PMID: 19614718.
- Posteraro B, Martucci R, La Sorda M, Fiori B, Sanglard D, De Carolis E, Florio AR, Fadda G, Sanguinetti M. Reliability of the Vitek 2 yeast susceptibility test for detection of in vitro resistance to fluconazole and voriconazole in clinical isolates of Candida albicans and Candida glabrata. J Clin Microbiol. 2009 Jun;47(6):1927-30. doi: 10.1128/JCM.02070-08. Epub 2009 Apr 29. PMID: 19403774; PMCID: PMC2691097.